|
Expert Views :: A Replaceable Hydrophobic Core Thomas Connor Bishop Historically, the estrogen receptor has held the leading role in the endocrine disrupter story (1), but it is only one member of a large class of ligand activated gene regulatory proteins, called the nuclear receptor superfamily (2). A number of x-ray crystallographic structures are publicly available for several members of this superfamily through the protein data bank. The images in this data bank constitute more than just a family photo album. The x-ray crystallographic structures provide the location and identification of every single atom that makes up a molecule, such as the above mentioned receptors. However, none of the x-ray structures, that have been determined to date, for this superfamily represent the structure of a complete receptor. The photo album, so to speak, only contains head-shots, hands, or feet — not a complete photo of an individual from the superfamily. Nonetheless, the x-ray structures provide valuable information. In particular, the ligand binding domain (say the feet) of this subfamily look very much alike in terms of their gross 3-Dimensional structure (see Figure 1 below). Yet, physiologically, it is known that each receptor is ligand activated by a different ligand and each receptor mediates a different gene regulatory response when activated (3). 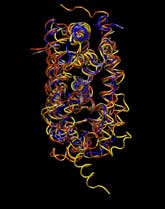
Since the intracellular environment is hydrophilic and since the residues are all connected together in a chain, intracellular proteins fold into a conformation that minimizes the exposure of hydrophobic residues and that maximizes the exposure of hydrophilic residues. This simple idea explains the existence of a "hydrophobic core" at the heart of most intracellular proteins. The steroid hormone receptors are unusual in that they have a replaceable hydrophobic core. By similar reasoning a hydrophobic compound that happens to find itself in a cell's hydrophilic interior is looking for a place to hide. It prefers a hydrophobic environment. Thus when a human or wildlife species is exposed to any of a number of natural or synthetic chemical compounds that are hydrophobic a situation is created at the molecular level, in which exogenous chemical agents compete with the body's natural chemical messengers to find a hydrophobic hiding place. Ultimately such competition between exogenous and endogenous chemicals can disrupt the endocrine system’s control over homeostasis, reproduction and development. Each of these processes is mediated in large part by activation and deactivation of the nuclear receptor proteins. Of course, the competition between exogenous and endogenous chemicals for a “hiding place” is not necessarily limited to just competitive binding to the receptor, but this certainly is an interesting possibility, and the subject of my research. Working in the Theoretical Molecular Biology Lab at the CBR with graduate student Kirk Yancy Williams, I am seeking to determine how various known endocrine disrupting compounds (7) change not only the structure but also the dynamics of the estrogen receptor. For this purpose we are utilizing different x-ray crystallographic structures of the estrogen receptor ligand binding domain (5, 6) (protein databank entries 1ERE, 3ERD, 1ERR, 3ERT) to create models of these ligand-receptor complexes. The resulting complexes are then subjected to all atom molecular dynamics simulations to determine an optimal ligand-receptor conformation and to investigate relationships between structure and dynamics. Analysis of the molecular dynamics enables us to identify the individual contribution from each atom of the ligand and of the receptor to the total free energy of binding. The free energy of binding is a thermodynamic property of the ligand-receptor complex that measures how energetically favorable the association of the ligand and the receptor is. From a practical perspective, these studies enable us to identify chemicals with an endocrine disruption potential, a priori. From a mechanistic perspective, knowledge of the effects of different ligands on the folding and the dynamics of the hormone receptors is a crucial component of our understanding of gene regulation by this class of proteins. References
|